07Structural and biochemical characterization of the M405S variant of Desulfovibrio vulgaris formate dehydrogenase
Molybdenum- or tungsten-dependent formate dehydrogenases have emerged as significant catalysts for the chemical reduction of CO2 to formate, with biotechnological applications envisaged in climate-change mitigation. The role of Met405 in the active site of Desulfovibrio vulgaris formate dehydrogenase AB (DvFdhAB) has remained elusive. However, its proximity to the metal site and the conformational change that it undergoes between the resting and active forms suggests a functional role. In this work, the M405S variant was engineered, which allowed the active-site geometry in the absence of methionine S(δ) interactions with the metal site to be revealed and the role of Met405 in catalysis to be probed. This variant displayed reduced activity in both formate oxidation and CO2 reduction, together with an increased sensitivity to oxygen inactivation.
Vilela-Alves, R. Rebelo Manuel, N. Pedrosa, I. A. Cardoso Pereira, M. J. Romão, and C. Mota,Structural and biochemical characterization of the M405S variant of Desulfovibrio vulgaris formate dehydrogenase, Acta Crystallogr F Struct Biol Commun, 2024, 80, 98-106.
Arsenite oxidase in complex with antimonite and arsenite oxyanions: Insights into the catalytic mechanism
Arsenic contamination of groundwater is among one of the biggest health threats affecting millions of people in the world. There is an urgent need for efficient arsenic biosensors where the use of arsenic metabolizing enzymes can be explored. In this work, we have solved four crystal structures of arsenite oxidase (Aio) in complex with arsenic and antimony oxyanions and the structures determined correspond to intermediate states of the enzymatic mechanism. These structural data were complemented with density-functional theory calculations providing a unique view of the molybdenum active site at different time points that, together with mutagenesis data, enabled to clarify the enzymatic mechanism and the molecular determinants for the oxidation of As(III) to the less toxic As(V) species.
- Engrola, M. A. S. Correia, C. Watson, C. C. Romão, L. F. Veiros, M. J. Romão, T. Santos-Silva, and J. M. Santini,Arsenite oxidase in complex with antimonite and arsenite oxyanions: Insights into the catalytic mechanism, J Biol Chem, 2023, 299, 105036.
Redox potentials elucidate the electron transfer pathway of NAD(+)-dependent formate dehydrogenases
Metal-dependent, nicotine adenine dinucleotide (NAD(+))-dependent formate dehydrogenases (FDHs) are complex metalloenzymes coupling biochemical transformations through intricate electron transfer pathways. Rhodobacter capsulatus FDH is a model enzyme for understanding coupled catalysis, in that reversible CO(2) reduction and formate oxidation are linked to a flavin mononuclotide (FMN)-bound diaphorase module via seven iron-sulfur (FeS) clusters as a dimer of heterotetramers. Catalysis occurs at a bis-metal-binding pterin (Mo) binding two molybdopterin guanine dinucleotides (bis-MGD), a protein-based Cys residue and a participatory sulfido ligand. Insights regarding the proposed electron transfer mechanism between the bis-MGD and the FMN have been complicated by the discovery that an alternative pathway might occur via intersubunit electron transfer between two [4Fe4S] clusters within electron transfer distance. To clarify this difference, the redox potentials of the bis-MGD and the FeS clusters were determined via redox titration by EPR spectroscopy. Redox potentials for the bis-MGD cofactor and five of the seven FeS clusters could be assigned. Furthermore, substitution of the active site residue Lys295 with Ala resulted in altered enzyme kinetics, primarily due to a more negative redox potential of the A1 [4Fe4S] cluster. Finally, characterization of the monomeric FdsGBAD heterotetramer exhibited slightly decreased formate oxidation activity and similar iron-sulfur clusters reduced relative to the dimeric heterotetramer. Comparison of the measured redox potentials relative to structurally defined FeS clusters support a mechanism by which electron transfer occurs within a heterotetrameric unit, with the interfacial [4Fe4S] cluster serving as a structural component toward the integrity of the heterodimeric structure to drive efficient catalysis.
R. Duffus, M. Gauglitz, C. Teutloff, and S. Leimkühler,Redox potentials elucidate the electron transfer pathway of NAD(+)-dependent formate dehydrogenases, J Inorg Biochem, 2024, 253, 112487.
potentials
Metal-Containing Formate Dehydrogenases, a Personal View
Mo/W-containing formate dehydrogenases FDH catalyzes the reversible oxidation of formate to carbon dioxide at their molybdenum or tungsten active sites. The metal-containing FDHs are members of the dimethylsulfoxide reductase family of mononuclear molybdenum cofactor Moco- or tungsten cofactor Wco-containing enzymes. In these enzymes, the active site in the oxidized state comprises a Mo or W atom present in the bis-Moco, which is coordinated by the two dithiolene groups from the two MGD moieties, a protein-derived SeCys or Cys, and a sixth ligand that is now accepted as being a sulfido group. SeCys-containing enzymes have a generally higher turnover number than Cys-containing enzymes. The analogous chemical properties of W and Mo, the similar active sites of W- and Mo-containing enzymes, and the fact that W can replace Mo in some enzymes have led to the conclusion that Mo- and W-containing FDHs have the same reaction mechanism. Details of the catalytic mechanism of metal-containing formate dehydrogenases are still not completely understood and have been discussed here.
Leimkühler,Metal-Containing Formate Dehydrogenases, a Personal View, Molecules, 2023, 28.
The Mechanism of Metal-Containing Formate Dehydrogenases Revisited: The Formation of Bicarbonate as Product Intermediate Provides Evidence for an Oxygen Atom Transfer Mechanism
Mo/W-containing formate dehydrogenases (FDH) catalyzed the reversible oxidation of formate to carbon dioxide at their molybdenum or tungsten active sites. While in the reaction of formate oxidation, the product is CO2, which exits the active site via a hydrophobic channel; bicarbonate is formed as the first intermediate during the reaction at the active site. Other than what has been previously reported, bicarbonate is formed after an oxygen atom transfer reaction, transferring the oxygen from water to formate and a subsequent proton-coupled electron transfer or hydride transfer reaction involving the sulfido ligand as acceptor.
Kumar, M. Khosraneh, S. S. M. Bandaru, C. Schulzke, and S. Leimkühler,The Mechanism of Metal-Containing Formate Dehydrogenases Revisited: The Formation of Bicarbonate as Product Intermediate Provides Evidence for an Oxygen Atom Transfer Mechanism, Molecules, 2023, 28.
07 Infrared Spectroscopy Elucidates the Inhibitor Binding Sites in a Metal-Dependent Formate Dehydrogenase
Biological carbon dioxide (CO2 ) reduction is an important step by which organisms form valuable energy-richer molecules required for further metabolic processes. The Mo-dependent formate dehydrogenase (FDH) from Rhodobacter capsulatus catalyzes reversible formate oxidation to CO2 at a bis-molybdopterin guanine dinucleotide (bis-MGD) cofactor. To elucidate potential substrate binding sites relevant for the mechanism, we studied herein the interaction with the inhibitory molecules azide and cyanate, which are isoelectronic to CO2 and charged as formate. We employed infrared (IR) spectroscopy in combination with density functional theory (DFT) and inhibition kinetics. One distinct inhibitory molecule was found to bind to either a non-competitive or a competitive binding site in the secondary coordination sphere of the active site. Site-directed mutagenesis of key amino acid residues in the vicinity of the bis-MGD cofactor revealed changes in both non-competitive and competitive binding, whereby the inhibitor is in case of the latter interaction presumably bound between the cofactor and the adjacent Arg587.
- Laun, B. R. Duffus, S. Wahlefeld, S. Katz, D. Belger, P. Hildebrandt, M. A. Mroginski, S. Leimkühler, and I. Zebger,Infrared Spectroscopy Elucidates the Inhibitor Binding Sites in a Metal-Dependent Formate Dehydrogenase, Chemistry, 2022, 28, e202201091.
07 Electrochemical Kinetics Support a Second Coordination Sphere Mechanism in Metal-Based Formate Dehydrogenase
Metal-based formate dehydrogenases are molybdenum or tungsten-dependent enzymes that catalyze the interconversion between formate and CO2 . According to the current consensus, the metal ion of the catalytic center in its active form is coordinated by 6 S (or 5 S and 1 Se) atoms, leaving no free coordination site to which formate could bind to the metal. Some authors have proposed that one of the active site ligands decoordinates during turnover to allow formate binding. Another proposal is that the oxidation of formate takes place in the second coordination sphere of the metal. Here, we have used electrochemical steady-state kinetics to elucidate the order of the steps in the catalytic cycle of two formate dehydrogenases. Our results strongly support the "second coordination sphere" hypothesis.
- Meneghello, A. Uzel, M. Broc, R. R. Manuel, A. Magalon, I. A. C. Pereira, C. Léger, A. Walburger, and V. Fourmond,Electrochemical Kinetics Support a Second Coordination Sphere Mechanism in Metal-Based Formate Dehydrogenase, Angew Chem Int Ed Engl, 2022.
FORMATE DEHYDROGENASE− Energetics for CO2 Reduction by Molybdenum-Containing Formate Dehydrogenase
The level of carbon dioxide in the atmosphere has increased in a dangerous way during the past century. Methods to decrease this level are therefore of high interest at present. Inspiration to do so in an efficient way could come from biological systems. Molybdenum-containing formate dehydrogenase Mo-FDH is one of the most interesting enzymes in this respect. For example, the reduction potential required is not very low. The normal reaction catalyzed by Mo-FDH is actually the opposite one of oxidizing formate to CO2. However, recent electrochemical studies have shown that the reaction can be reversed by a moderate lowering of the reduction potential. The goal of the present study has been to study the full mechanism of Mo-FDH, particularly in the most interesting direction of reducing CO2, which has not been done before. The methods used are the same as those that have been shown to give excellent results for redox enzymes in all cases they have been tested. The results obtained for Mo-FDH are also in excellent agreement with the experimental results.
P. E. M. Siegbahn,Energetics for CO2 Reduction by Molybdenum-Containing Formate Dehydrogenase, J Phys Chem B, 2022, 126, 1728-1733.
Formate Dehydrogenases Reduce CO2 Rather than HCO3-: An Electrochemical Demonstration
Mo/W formate dehydrogenases catalyze the reversible reduction of CO2 species to formate. It is thought that the substrate is CO2 and not a hydrated species like HCO3-, but there is still no indisputable evidence for this, in spite of the extreme importance of the nature of the substrate for mechanistic studies. We devised a simple electrochemical method to definitively demonstrate that the substrate of formate dehydrogenases is indeed CO2.
M. Meneghello, A. R. Oliveira, A. Jacq-Bailly, I. A. C. Pereira, C. Leger, and V. Fourmond,Formate Dehydrogenases Reduce CO2 Rather than HCO3-: An Electrochemical Demonstration, Angewandte Chemie-International Edition.2021. https://doi.org/10.1002/ange.202101167
FORMATE DEHYDROGENASE
Classification and enzyme kinetics of formate dehydrogenases for biomanufacturing via CO2 utilization
The reversible interconversion of formate (HCOO-) and carbon dioxide (CO2) is catalyzed by formate dehydrogenase (FDH, EC 1.17.1.9). This enzyme can be used as a first step in the utilization of CO2 as carbon substrate for production of high-in-demand chemicals. However, comparison and categorization of the very diverse group of FDH enzymes has received only limited attention. With specific emphasis on FDH catalyzed CO2 reduction to HCOO-, we present a novel classification scheme for FDHs based on protein sequence alignment and gene organization analysis. We show that prokaryotic FDHs can be neatly divided into six meaningful sub-types. These sub-types are discussed in the context of overall structural composition, phylogeny of the gene segment organization, metabolic role, and catalytic properties of the enzymes. Based on the available literature, the influence of electron donor choice on the efficacy of FDH catalyzed CO2 reduction is quantified and compared. This analysis shows that methyl viologen and hydrogen are several times more potent than NADH as electron donors. Hence, the new FDH classification scheme and the electron donor analysis provide an improved base for developing FDH-facilitated CO2 reduction as a viable step in the utilization of CO2 as carbon source for green production of chemicals.
C. F. Nielsen, L. Lange, and A. S. Meyer,Classification and enzyme kinetics of formate dehydrogenases for biomanufacturing via CO2 utilization, Biotechnology Advances, 2019, 37.
Formate dehydrogenase
Deconvolution of reduction potentials of formate dehydrogenase from Cupriavidus necator
The formate dehydrogenase enzyme from Cupriavidus necator (FdsABG) carries out the two-electron oxidation of formate to CO2, but is also capable of reducing CO2 back to formate, a potential biofuel. FdsABG is a heterotrimeric enzyme that performs this transformation using nine redox-active cofactors: a bis(molybdopterin guanine dinucleotide) (bis-MGD) at the active site coupled to seven iron-sulfur clusters, and one equivalent of flavin mononucleotide (FMN). To better understand the pathway of electron flow in FdsABG, the reduction potentials of the various cofactors were examined through direct electrochemistry. Given the redundancy of cofactors, a truncated form of the FdsA subunit was developed that possesses only the bis-MGD active site and a singular [4Fe-4S] cluster. Electrochemical characterization of FdsABG compared to truncated FdsA shows that the measured reduction potentials are remarkably similar despite the truncation with two observable features at - 265 mV and - 455 mV vs SHE, indicating that the voltammetry of the truncated enzyme is representative of the reduction potentials of the intact heterotrimer. By producing truncated FdsA without the necessary maturation factors required for bis-MGD insertion, a form of the truncated FdsA that possesses only the [4Fe-4S] was produced, which gives a single voltammetric feature at - 525 mV, allowing the contributions of the molybdenum cofactor to be associated with the observed feature at - 265 mV. This method allowed for the deconvolution of reduction potentials for an enzyme with highly complex cofactor content to know more about the thermodynamic landscape of catalysis.
L. M. Walker, B. Li, D. Niks, R. Hille, and S. J. Elliott,Deconvolution of reduction potentials of formate dehydrogenase from Cupriavidus necator, Journal of biological inorganic chemistry : JBIC : a publication of the Society of Biological Inorganic Chemistry, 2019, 24, 889-898.
Formate dehydrogenase
Deconvolution of reduction potentials of formate dehydrogenase from Cupriavidus necator
The formate dehydrogenase enzyme from Cupriavidus necator (FdsABG) carries out the two-electron oxidation of formate to CO2, but is also capable of reducing CO2 back to formate, a potential biofuel. FdsABG is a heterotrimeric enzyme that performs this transformation using nine redox-active cofactors: a bis(molybdopterin guanine dinucleotide) (bis-MGD) at the active site coupled to seven iron-sulfur clusters, and one equivalent of flavin mononucleotide (FMN). To better understand the pathway of electron flow in FdsABG, the reduction potentials of the various cofactors were examined through direct electrochemistry. Given the redundancy of cofactors, a truncated form of the FdsA subunit was developed that possesses only the bis-MGD active site and a singular [4Fe-4S] cluster. Electrochemical characterization of FdsABG compared to truncated FdsA shows that the measured reduction potentials are remarkably similar despite the truncation with two observable features at - 265 mV and - 455 mV vs SHE, indicating that the voltammetry of the truncated enzyme is representative of the reduction potentials of the intact heterotrimer. By producing truncated FdsA without the necessary maturation factors required for bis-MGD insertion, a form of the truncated FdsA that possesses only the [4Fe-4S] was produced, which gives a single voltammetric feature at - 525 mV, allowing the contributions of the molybdenum cofactor to be associated with the observed feature at - 265 mV. This method allowed for the deconvolution of reduction potentials for an enzyme with highly complex cofactor content to know more about the thermodynamic landscape of catalysis.
L. M. Walker, B. Li, D. Niks, R. Hille, and S. J. Elliott,Deconvolution of reduction potentials of formate dehydrogenase from Cupriavidus necator, Journal of biological inorganic chemistry : JBIC : a publication of the Society of Biological Inorganic Chemistry, 2019.
Molybdenum- and tungsten-containing formate dehydrogenases and formylmethanofuran dehydrogenases: Structure, mechanism, and cofactor insertion
An overview is provided of the molybdenum- and tungsten-containing enzymes that catalyze the interconversion of formate and CO2 , focusing on common structural and mechanistic themes, as well as a consideration of the manner in which the mature Mo- or W-containing cofactor is inserted into apoprotein.
D. Niks, and R. Hille,Molybdenum- and tungsten-containing formate dehydrogenases and formylmethanofuran dehydrogenases: Structure, mechanism, and cofactor insertion, Protein science : a publication of the Protein Society, 2019, 28, 111-122.
Efficient reduction of CO2 by the molybdenum-containing formate dehydrogenase from Cupriavidus necator (Ralstonia eutropha)
The ability of the FdsABG formate dehydrogenase from Cupriavidus necator (formerly known as Ralstonia eutropha) to catalyze the reverse of the physiological reaction, the reduction of CO2 to formate utilizing NADH as electron donor, has been investigated. Contrary to previous studies of this enzyme, we demonstrate that it is in fact effective in catalyzing the reverse reaction with a k(cat) of 11 +/- 0.4 s(-1). We also quantify the stoichiometric accumulation of formic acid as the product of the reaction and demonstrate that the observed kinetic parameters for catalysis in the forward and reverse reactions are thermodynamically consistent, complying with the expected Haldane relationships. Finally, we demonstrate the reaction conditions necessary for gauging the ability of a given formate dehydrogenase or other CO2-utilizing enzyme to catalyze the reverse direction to avoid false negative results. In conjunction with our earlier studies on the reaction mechanism of this enzyme and on the basis of the present work, we conclude that all molybdenum- and tungsten-containing formate dehydrogenases and related enzymes likely operate via a simple hydride transfer mechanism and are effective in catalyzing the reversible interconversion of CO2 and formate under the appropriate experimental conditions.
Yu, X. J., Niks, D., Mulchandani, A., and Hille, R.,Efficient reduction of CO2 by the molybdenum-containing formate dehydrogenase from Cupriavidus necator (Ralstonia eutropha), Journal of Biological Chemistry, 2017, 292, 16872-16879
Formate dehydrogenase
Molybdenum and tungsten-containing formate dehydrogenases: Aiming to inspire a catalyst for carbon dioxide utilization
The global energy demand and the present high dependence on fossil fuels have caused an unprecedented increase in the Earth's atmosphere carbon dioxide concentration. Its exponential and uncontrollable rise is responsible for large and unpredictable impacts on the world climate and for ocean acidification, thus, being a major concern for the ecosystems and human's daily life. On the other hand, the carbon dioxide abundance and low cost make it an interesting source for the production of chemical feedstocks and fuels. Yet, the thermodynamic and kinetic stability of the carbon dioxide molecule makes its utilization a laboratorial/industrially challenging task. In this Review, we propose to use the molybdenum and tungsten-containing formate dehydrogenase (FDH) enzymes as a model to understand the mechanistic strategies and key chemical features needed to reduce carbon dioxide to formate. We will highlight the present knowledge about the structure of FDHs, with particular emphasis on active site features, reaction mechanism and ability to reduce carbon dioxide to formate. The information gathered aims to inspire the development of new efficient (bio)catalysts for the atmospheric carbon dioxide utilization, to produce energy and chemical feedstocks, while reducing an important environmental pollutant. (C) 2016 Elsevier B.V. All rights reserved.
Maia, L. B., Moura, I., and Moura, J. J. G., Molybdenum and tungsten-containing formate dehydrogenases: Aiming to inspire a catalyst for carbon dioxide utilization, Inorganica Chimica Acta, 2017, 455, 350-363.
Formate dehydrogenase
Molybdenum- and tungsten-containing formate dehydrogenases and formylmethanofuran dehydrogenases: Structure, mechanism and cofactor insertion
An overview is provided of the molybdenum- and tungsten-containing enzymes that catalyze the interconversion of formate and CO2 , focusing on common structural and mechanistic themes, as well as a consideration of the manner in which the mature Mo- or W-containing cofactor is inserted into apoprotein. This article is protected by copyright. All rights reserved.
D. Niks, and R. Hille, Molybdenum- and tungsten-containing formate dehydrogenases and formylmethanofuran dehydrogenases: Structure, mechanism and cofactor insertion, Protein Sci, 2018.
Spectroscopic and kinetic properties of the molybdenum-containing, NAD(+) - dependent formate dehydrogenase from ralstonia eutropha
We have examined the rapid reaction kinetics and spectroscopic properties of the molybdenum-containing, NAD(+) -dependent FdsABG formate dehydrogenase from Ralstonia eutropha. We confirm previous steady-state studies of the enzyme and extend its characterization to a rapid kinetic study of the reductive half-reaction (the reaction of formate with oxidized enzyme). We have also characterized the electron paramagnetic resonance signal of the molybdenum center in its Mo-V state and demonstrated the direct transfer of the substrate C alpha hydrogen to the molybdenum center in the course of the reaction. Varying temperature, microwave power, and level of enzyme reduction, we are able to clearly identify the electron paramagnetic resonance signals for four of the iron/sulfur clusters of the enzyme and find suggestive evidence for two others; we observe a magnetic interaction between the molybdenum center and one of the iron/sulfur centers, permitting assignment of this signal to a specific iron/sulfur cluster in the enzyme. In light of recent advances in our understanding of the structure of the molybdenum center, we propose a reaction mechanism involving direct hydride transfer from formate to a molybdenum-sulfur group of the molybdenum center.
Niks, D., Duvvuru, J., Escalona, M., and Hille, R.,Spectroscopic and Kinetic Properties of the Molybdenum-containing, NAD(+) - dependent Formate Dehydrogenase from Ralstonia eutropha, Journal of Biological Chemistry, 2016, 291, 1162-1174.
Formate and nitrate. The sulfur shift: An activation mechanism for periplasmic nitrate reductase and formate dehydrogenase
The sulfur-shift mechanism is characterized by the displacement of the coordinating cysteine (in Nap or SeCys in Fdh) side chain to a second shell of the Mo-coordination sphere. This rearrangement enables a free coordination position for substrate binding to Mo ion and provides an efficient mechanism to maintaining a constant coordination number throughout the entire catalytic pathway. This type of mechanism is very similar to the carboxylate shift observed in other enzymes, and it has been recently detected by experimental means.
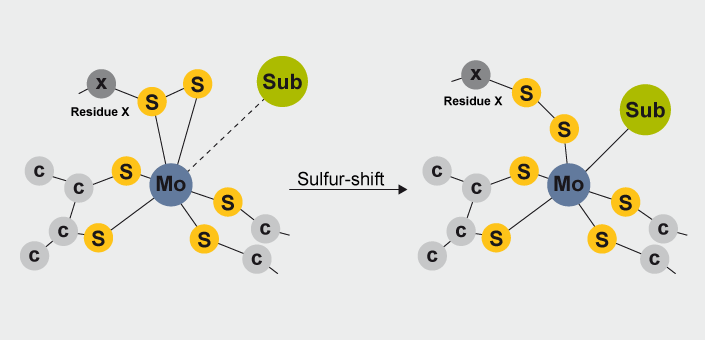
A structural rearrangement known as sulfur shift occurs in some Mo-containing enzymes of the DMSO reductase family. This mechanism is characterized by the displacement of a coordinating cysteine thiol (or SeCys in Fdh) from the first to the second shell of the Mo-coordination sphere metal. The hexa-coordinated Mo ion found in the as-isolated state cannot bind directly any exogenous ligand (substrate or inhibitors), while the penta-coordinated ion, attained upon sulfur shift, has a free binding site for direct coordination of the substrate. This rearrangement provides an efficient mechanism to keep a constant coordination number throughout an entire catalytic pathway. This mechanism is very similar to the carboxylate shift observed in Zn-dependent enzymes, and it has been recently detected by experimental means. In the present paper, we calculated the geometries and energies involved in the sulfur-shift mechanism using QM-methods (M06/(6-311++G(3df,2pd),SDD)//B3LYP/(6-31G(d),SDD)). The results indicated that the sulfur-shift mechanism provides an efficient way to enable the metal ion for substrate coordination.
Cerqueira, N. M., Fernandes, P. A., Gonzalez, P. J., Moura, J. J., and Ramos, M. J. The Sulfur Shift: An Activation Mechanism for Periplasmic Nitrate Reductase and Formate Dehydrogenase. Inorg. Chem., 2013, 52 , 10766–10772.
Formate and nitrate. periplasmic nitrate reductase and formate dehydrogenase: Similar molecular architectures with very different enzymatic activities
It is remarkable how nature has been able to construct enzymes that, despite sharing many similarities, have simple but key differences that tune them for completely different functions in living cells. Periplasmic nitrate reductase (Nap) and formate dehydrogenase (Fdh) from the DMSOr family are representative examples of this. Both enzymes share almost identical three-dimensional protein foldings and active sites, in terms of coordination number, geometry and nature of the ligands. The substrates of both enzymes (nitrate and formate) are polyatomic anions that also share similar charge and stereochemistry. In terms of the catalytic mechanism, both enzymes have a common activation mechanism (the sulfur-shift mechanism) that ensures a constant coordination number around the metal ion during the catalytic cycle. In spite of these similarities, they catalyze very different reactions: Nap abstracts an oxygen atom from nitrate releasing nitrite, whereas FdH catalyzes a hydrogen atom transfer from formate and releases carbon dioxide. In this Account, a critical analysis of structure, function, and catalytic mechanism of the molybdenum enzymes periplasmic nitrate reductase (Nap) and formate dehydrogenase (Fdh) is presented. We conclude that the main structural driving force that dictates the type of reaction, catalyzed by each enzyme, is a key difference on one active site residue that is located in the top region of the active sites of both enzymes. In both enzymes, the active site is centered on the metal ion of the cofactor (Mo in Nap and Mo or W in Fdh) that is coordinated by four sulfur atoms from two pyranopterin guanosine dinucleotide (PGD) molecules and by a sulfido. However, while in Nap there is a Cys directly coordinated to the Mo ion, in FdH there is a SeCys instead. In Fdh there is also an important His that interacts very closely with the SeCys, whereas in Nap the same position is occupied by a Met. The role of Cys in Nap and SeCys in FdH is similar in both enzymes; however, Met and His have different roles. His participates directly on catalysis, and it is therefore detrimental for the catalytic cycle of FdH. Met only participates in substrate binding. We concluded that this small but key difference dictates the type of reaction that is catalyzed by each enzyme. In addition, it allows explaining why formate can bind in the Nap active site in the same way as the natural substrate (nitrate), but the reaction becomes stalled afterward.
Cerqueira, N. M., Gonzalez, P. J., Fernandes, P. A., Moura, J. J., and Ramos, M. J., Periplasmic Nitrate Reductase and Formate Dehydrogenase: Similar Molecular Architectures with Very Different Enzymatic Activities, Accounts of chemical research, 2015.
[Periplasmic: The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria. Cytoplasm is the fluid that fills a cell. Wikipedia.]
Assembly and catalysis of molybdenum or tungsten-containing formate dehydrogenases from bacteria
The global carbon cycle depends on the biological transformations of C1 compounds, which include the reductive incorporation of CO2 into organic molecules (e.g. in photosynthesis and other autotrophic pathways), in addition to the production of CO2 from formate, a reaction that is catalyzed by formate dehydrogenases (FDHs). FDHs catalyze, in general, the oxidation of formate to CO2 and H+. However, selected enzymes were identified to act as CO2 reductases, which are able to reduce CO2 to formate under physiological conditions. This reaction is of interest for the generation of formate as a convenient storage form of H2 for future applications. Cofactor-containing FDHs are found in anaerobic bacteria and archaea, in addition to facultative anaerobic or aerobic bacteria. These enzymes are highly diverse and employ different cofactors such as the molybdenum cofactor (Moco), FeS clusters and flavins, or cytochromes. Some enzymes include tungsten (W) in place of molybdenum (Mo) at the active site. For catalytic activity, a selenocysteine (SeCys) or cysteine (Cys) ligand at the Mo atom in the active site is essential for the reaction. This review will focus on the characterization of Mo- and W-containing FDHs from bacteria, their active site structure, subunit compositions and its proposed catalytic mechanism. We will give an overview on the different mechanisms of substrate conversion available so far, in addition to providing an outlook on bio-applications of FDHs. This article is part of a Special Issue entitled: Cofactor-dependent proteins: evolution, chemical diversity and bio-applications.
Hartmann, T., Schwanhold, N., and Leimkuhler, S.,Assembly and catalysis of molybdenum or tungsten-containing formate dehydrogenases from bacteria, Biochimica et biophysica acta, 2015, 1854, 1090-100.
Mechanism
The prokaryotic formate metabolism is considerably diversified. Prokaryotes use formate in the C1 metabolism, but also evolved to exploit the low reduction potential of formate to derive energy, by coupling its oxidation to the reduction of numerous electron acceptors. To fulfil these varied physiological roles, different types of formate dehydrogenase (FDH) enzymes have evolved to catalyse the reversible 2-electron oxidation of formate to carbon dioxide. This review will highlight our present knowledge about the diverse physiological roles of FDH in prokaryotes, their modular structural organisation and active site structures and the mechanistic strategies followed to accomplish the formate oxidation. In addition, the ability of FDH to catalyse the reverse reaction of carbon dioxide reduction, a potentially relevant reaction for carbon dioxide sequestration, will also be addressed.
Maia LB, Moura JJ, Moura I. Molybdenum and tungsten-dependent formate dehydrogenases. J Biol Inorg Chem. 2015 Mar;20(2):287-309. doi: 10.1007/s00775-014-1218-2. Epub 2014 Dec 5.
Escherichia coli can perform two modes of formate metabolism. Under respiratory conditions, two periplasmically-located formate dehydrogenase isoenzymes couple formate oxidation to the generation of a transmembrane electrochemical gradient; and under fermentative conditions a third cytoplasmic isoenzyme is involved in the disproportionation of formate to CO2 and H2. The respiratory formate dehydrogenases are redox enzymes that comprise three subunits: a molybdenum cofactor- and FeS cluster-containing catalytic subunit; an electron-transferring ferredoxin; and a membrane-integral cytochrome b. The catalytic subunit and its ferredoxin partner are targeted to the periplasm as a complex by the twin-arginine transport (Tat) pathway. Biosynthesis of these enzymes is under control of an accessory protein, FdhE. E. coli FdhE interacts with the catalytic subunits of the respiratory formate dehydrogenases. Purification of recombinant FdhE demonstrates the protein is an iron-binding rubredoxin that can adopt monomeric and homodimeric forms. Bacterial two-hybrid analysis suggests the homodimer form of FdhE is stabilized by anaerobiosis. Site-directed mutagenesis shows that conserved cysteine motifs are essential for the physiological activity of the FdhE protein and are also involved in iron ligation
Luke, I., Butland, G., Moore, K., Buchanan, G., Lyall, V., Fairhurst, S., Greenblatt, J., Emili, A., Palmer, T., and Sargent, F., Biosynthesis of the respiratory formate dehydrogenases from Escherichia coli: characterization of the FdhE protein, Archives of Microbiology, 2008, 190, 685-696.
The mechanism of formate oxidation by metal-dependent formate dehydrogenases
Metal-dependent formate dehydrogenases (Fdh) from prokaryotic organisms are members of the dimethyl sulfoxide reductase family of mononuclear molybdenum-containing and tungsten-containing enzymes. Fdhs catalyze the oxidation of the formate anion to carbon dioxide in a redox reaction that involves the transfer of two electrons from the substrate [formate] to the active site.
The active site in the oxidized state comprises a hexacoordinated molybdenum or tungsten ion in a distorted trigonal prismatic geometry. Using this structural model, we calculated the catalytic mechanism of Fdh through density functional theory tools.
The simulated mechanism was correlated with the experimental kinetic properties of three different Fdhs isolated from three different Desulfovibrio species.
Our studies indicate that the C-H bond break is an event involved in the rate-limiting step of the catalytic cycle.
The role in catalysis of conserved amino acid residues involved in metal coordination and near the metal active site is discussed on the basis of experimental and theoretical results
Mota, Cristiano S., Rivas, Maria G., Brondino, Carlos D., Moura, Isabel, Moura, Jose J. G., Gonzalez, Pablo J., and Cerqueira, Nuno M. F. S., The mechanism of formate oxidation by metal-dependent formate dehydrogenases, Journal of Biological Inorganic Chemistry, 2011, 16, 1255-1268.
Clostridium carboxidivorans strain P7T recombinant formate dehydrogenase catalyzes reduction of CO2 to formate
Recombinant formate dehydrogenase from the acetogen Clostridium carboxidivorans strain P7(T), expressed in Escherichia coli, shows particular activity towards NADH-dependent carbon dioxide reduction to formate due to the relative binding affinities of the substrates and products. The enzyme retains activity over 2 days at 4 degrees C under oxic conditions
Alissandratos, A., Kim, H. K., Matthews, H., Hennessy, J. E., Philbrook, A., and Easton, C. J., Clostridium carboxidivorans Strain P7T Recombinant Formate Dehydrogenase Catalyzes Reduction of CO2 to Formate, Applied and Environmental Microbiology, 2013, 79, 741-744.
Periplasmic nitrate reductases and formate dehydrogenases: Biological control of the chemical properties of Mo and W for fine tuning of reactivity, substrate specificity and metabolic role
Mo- and W-enzymes are widely distributed in biology as they can be found in all domains of life. They perform key roles in several metabolic pathways catalyzing important reactions of the biogeochemical cycles of the more abundant elements of the earth. These reactions are usually redox processes involving the transfer of an atom from the substrate to the metal ion or vice versa. The Mo or W reactivity and specificity toward a substrate is determined by the polypeptide chain of the enzyme, which tunes the chemical properties of the metal ion. Two enzymes sharing almost identical active sites but catalyzing very different reactions are periplasmic nitrate reductase and formate dehydrogenase from bacteria. They represent a good example of how key changes in the amino acid sequence tune the properties of an enzyme. In order to analyze the chemistry of Mo and W in these enzymes, structural, kinetic and spectroscopic data are reviewed, along with the role of these enzymes in cell metabolism. In addition, the features that govern selectivity of metal uptake into the cell and Mo/W-cofactor biosynthesis are revised. (C) 2012 Published by Elsevier B.V
Gonzalez, P. J., Rivas, M. G., Mota, C. S., Brondino, C. D., Moura, I., and Moura, J. J. G., Periplasmic nitrate reductases and formate dehydrogenases: Biological control of the chemical properties of Mo and W for fine tuning of reactivity, substrate specificity and metabolic role, Coordination Chemistry Reviews, 2013, 257, 315-331.

