Molybdopterin synthase
Molybdopterin cofactor - biosynthesis
Background: Localization and identification of interaction partners of two splice variants of the human 3-mercaptopyruvate sulfurtransferase TUM1.
Results: We show that TUM1 interacts with proteins involved in Moco and FeS cluster biosynthesis.
Conclusion: Human TUM1 is a dual localized protein in the cytosol and mitochondria with distinct roles in sulfur transfer and interaction partners.
Significance: The study contributes to the sulfur transfer pathway for the biosynthesis of sulfur-containing biofactors. The human tRNA thiouridine modification protein (TUM1), also designated as 3-mercaptopyruvate sulfurtransferase (MPST), has been implicated in a wide range of physiological processes in the cell. The roles range from an involvement in thiolation of cytosolic tRNAs to the generation of H2S as signaling molecule both in mitochondria and the cytosol. TUM1 is a member of the sulfurtransferase family and catalyzes the conversion of 3-mercaptopyruvate to pyruvate and protein-bound persulfide. Here, we purified and characterized two novel TUM1 splice variants, designated as TUM1-Iso1 and TUM1-Iso2. The purified proteins showed similar kinetic behavior and comparable pH and temperature dependence. Cellular localization studies, however, showed a different localization pattern between the isoforms. TUM1-Iso1 is exclusively localized in the cytosol, whereas TUM1-Iso2 showed a dual localization both in the cytosol and mitochondria. Interaction studies were performed with the isoforms both in vitro using the purified proteins and in vivo by fluorescence analysis in human cells, using the split-EGFP system. The studies showed that TUM1 interacts with the l-cysteine desulfurase NFS1 and the rhodanese-like protein MOCS3, suggesting a dual function of TUM1 both in sulfur transfer for the biosynthesis of the molybdenum cofactor, and for the thiolation of tRNA. Our studies point to distinct roles of each TUM1 isoform in the sulfur transfer processes in the cell, with different compartmentalization of the two splice variants of TUM1.
Frasdorf, B., Radon, C., and Leimkuhler, S., Characterization and Interaction Studies of Two Isoforms of the Dual Localized 3-Mercaptopyruvate Sulfurtransferase TUM1 from Humans, Journal of Biological Chemistry, 2014, 289, 34543-34556.
Enzyme Cofactor
The biosynthesis of the molybdenum cofactors (Moco) is an ancient, ubiquitous, and highly conserved pathway leading to the biochemical activation of molybdenum. Moco is the essential component of a group of redox enzymes, which are diverse in terms of their phylogenetic distribution and their architectures, both at the overall level and in their catalytic geometry. A wide variety of transformations are catalyzed by these enzymes at carbon, sulfur and nitrogen atoms, which include the transfer of an oxo group or two electrons to or from the substrate. More than 50 molybdoenzymes were identified to date. In all molybdoenzymes except nitrogenase, molybdenum is coordinated to a dithiolene group on the 6-alkyl side chain of a pterin called molybdopterin (MPT). The biosynthesis of Moco can be divided into three general steps, with a fourth one present only in bacteria and archaea: (1) formation of the cyclic pyranopterin monophosphate, (2) formation of MPT, (3) insertion of molybdenum into molybdopterin to form Moco, and (4) additional modification of Moco in bacteria with the attachment of a nucleotide to the phosphate group of MPT, forming the dinucleotide variant of Moco. This review will focus on the biosynthesis of Moco in bacteria, humans and plants.
Mendel RR(1), Leimkühler S. The biosynthesis of the molybdenum cofactors. J Biol Inorg Chem. 2015 Mar;20(2):337-47. doi: 10.1007/s00775-014-1173-y. Epub 2014 Jul 1.
Molybdopterin is inserted into xanthine dehydrogenase only after molybdenum chelation, and both metal chelation and molybdopterin insertion can occur only under high molybdate concentrations. In the biosynthesis of the molybdenum cofactor the biosynthesis of molybdopterin and molybdopterin guanine dinucleotide are split at a stage when the molybdenun atom is added to molybdopterin.
Leimkuhler, S., Angermuller, S., Schwarz, G., Mendel, R.R., Klipp,W., Activity of the molybdopterin-containing xanthine dehydrogenase of Rhodobacter capsulatus can be restored by high molybdenum concentrations in a moeA mutant defective in molybdenum cofactor biosynthesis, Journal Of Bacteriology , 1999, 181 , 19, 5930-5939.
Molybdenum cofactor (Moco) biosynthesis is an evolutionarily conserved pathway present in eubacteria, archaea and eukaryotes, including humans. Genetic deficiencies of enzymes involved in Moco biosynthesis in humans lead to a severe and usually fatal disease. Moco contains a tricyclic pyranopterin, termed molybdopterin, that bears the cis-dithiolene group responsible for molybdenum ligation. The dithiolene group of molybdopterin is generated by molybdopterin synthase, which consists of a large and small subunits. The 1.45 Angstrom resolution crystal structure of molybdopterin synthase reveals a heterotetrameric protein in which the C-terminus of each small subunit is inserted into a large subunit to form the active site. In the activated form of the enzyme this C-terminus is present as a thiocarboxylate. In the structure of a covalent complex of molybdopterin synthase, an isopeptide bond is present between the C-terminus of the small subunit and a Lys side chain in the large subunit. The strong structural similarity between the small subunit of molybdopterin synthase and ubiquitin provides evidence for the evolutionary antecedence of the Moco biosynthetic pathway to the ubiquitin dependent protein degradation pathway.
Rudolph, M.J., Wuebbens, M. M., Rajagopalan, K. V., and Schindelin, H., Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation, Nature Structural Biology, 2001, 8, 42-46.
See also
Schrag, J.D., Huang, W. J., Sivaraman, J., Smith, C., Plamondon, J., Larocque, R., Matte, A., and Cygler, M., The crystal structure of Escherichia coli MoeA, a protein from the molybdopterin synthesis pathway, Journal of Molecular Biology , 2001, 310, 419-431.
Molybdopterin is a pyranopterin with a unique dithiolene group coordinating molybdenum or tungsten in all molybdenum- and tungsten-enzymes except nitrogenase. In Escherichia coli, molybdopterin is formed by incorporation of two sulfur atoms into a precursor, which is catalyzed by the molybdopterin synthase enzyme. A two-step reaction of molybdopterin synthesis is proposed where the dithiolene is generated by two thiocarboxylates derived from a single tetrameric molybdopterin synthase.
Gutzke, G., Fischer, B., Mendel, R. R., and Schwarz, G., Thiocarboxylation of molybdopterin synthase provides evidence for the mechanism of dithiolene formation in metal-binding pterins, Journal of Biological Chemistry, 2001, 276, 36268-36274.
See also
Leimkuhler, S., Wuebbens, M. M., and Rajagopalan, K. V., Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor, Journal of Biological Chemistry, 2001, 276, 34695-34701.
Krepinsky, K. and Leimkuhler, S., Site-directed mutagenesis of the active site loop of the rhodanese-like domain of the human molybdopterin synthase sulfurase MOCS3 - Major differences in substrate specificity between eukaryotic and bacterial homologs, Febs Journal, 2007, 274, 2778-2787.
Regulski, E.E., Moy, R. H., Weinberg, Z., Barrick, J. E., Yao, Z., Ruzzo, W. L., and Breaker, R. R., A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism, Molecular Microbiology, 2008, 68, 918-932.
Biosynthesis of molybdenum and tungsten enzymes
See Mo transport.
Bevers, L. E., Hagedoorn, P.L., Santamaria-Araujo, J.A., Magalon, A., Hagen, W.R., Schwarz, G., BIOCHEMISTRY, 47, 949-956, 2008.
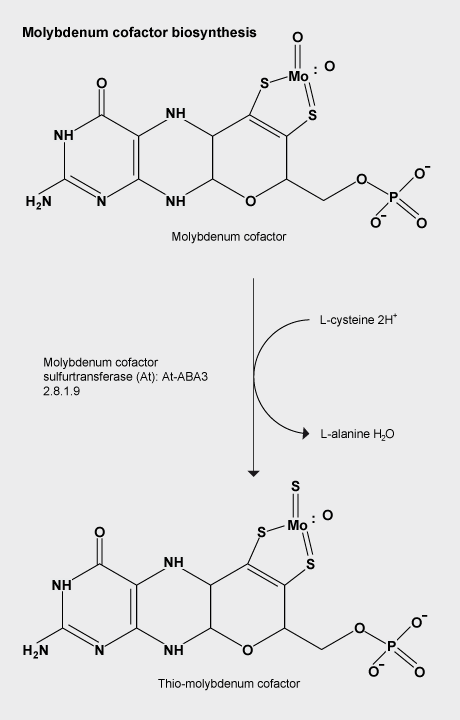
Molybdenum cofactor sulfuration
In almost all biological life forms, molybdenum and tungsten are coordinated by molybdopterin (MPT), a tricyclic pyranopterin containing a cis-dithiolene group. Molybdenum and the pterin moiety form the redox reactive molybdenum cofactor (Moco). Mutations in patients with deficiencies in Moco biosynthesis usually occur in the enzymes catalyzing the first and second steps of biosynthesis, leading to the formation of: precursor Z and MPT, respectively. The conversion of the sulfur- and metal-free precursor Z to MPT by MPT synthase involves sulfur atom transfer from a C-terminal MoaD thiocarboxylate to the C-1' and C-2' positions of precursor Z. The crystal structures of non-thiocarboxylated MPT synthase from Staphylococcus aureus in its apo form and in complex with precursor Z. A is reported.
Daniels, J. N., Wuebbens, M. M., Rajagopalan, K. V., and Schindelin, H., Crystal structure of a molybdopterin synthase-precursor Z complex: Insight into its sulfur transfer mechanism and its role in molybdenum cofactor deficiency, Biochemistry, 2008, 47,615-626, 3315.
Wollers, S., Heidenreich, T., Zarepour, M., Zachmann, D., Kraft, C., Zhao, Y. D., Mendel, R. R., and Bittner, F., Binding of sulfurated molybdenum cofactor to the C-terminal domain of ABA3 from Arabidopsis thaliana provides insight into the mechanism of molybdenum cofactor sulfuration, Journal of Biological Chemistry, 2008, 283, 9642-9650.
Molybdopterin co-factor coordination chemistry review
Dithiolenes in natural systems are ligands that bind molybdenum or tungsten at the catalytic centre of enzymes which catalyse the transfer of an oxygen atom to or from the substrate: e.g. the sulfite oxidases, sulfite to sulfate, and the nitrate reductases, nitrate to nitrite. The catalytic centres have one or two molybdopterin (MPT) cofactors bound to a mononuclear metal centre via their dithiolene group. The review covers the biosynthesis of MPT, its role in the function of the oxotransferase enzymes and the coordination chemistry that has been stimulated by the present knowledge of the nature and function of the catalytic centres of these enzymes.
Hine, F. J., Taylor, A. J., and Garner, C. D., Dithiolene complexes and the nature of molybdopterin, Coordination Chemistry Reviews, 2010, 254, 1570-1579.
FeMo-co precursor
George, S.J., Igarashi, R. Y., Xiao, Y., Hernandez, J. A., Demuez, M., Zhao, D., Yoda, Y., Ludden, P. W., Rubio, L. M., and Cramer, S. P., Extended X-ray absorption fine structure and nuclear resonance vibrational Spectroscopy reveal that NifB-co, a FeMo-co precursor, comprises a 6Fe core with an interstitial light atom, Journal of the American Chemical Society, 2008, 130, 5673-5680.
Inhibitor
Johannes, J., Unciuleac, M. C., Friedrich, T., Warkentin, E., Ermler, U., and Boll, M., Inhibitors of the molybdenum cofactor containing 4-Hydroxybenzoyl-CoA reductase, Biochemistry, 2008, 47, 4964-4972.
Sulfuration
The Moco (molybdenum cofactor) sulfurase ABA3 from Arabidopsis thaliana catalyses the sulfuration of the Moco of aldehyde oxidase and xanthine oxidoreductase, which represents the final activation step of these enzymes.
ABA3 consists of an N-terminal NifS-like domain that exhibits L-cysteine desulfurase activity and a C-terminal domain that binds sulfurated Moco.
The strictly conserved Cys(430) in the NifS-like domain binds a persulfide intermediate, which is abstracted from the substrate L-cysteine and finally needs to be transferred to the Moco of aldehyde oxidase and xanthine oxidoreductase.
In addition to Cys(430), another eight cysteine residues are located in the NifS-like domain, with two of them being highly conserved among Moco sulfurase proteins and, at the same time, being in close proximity to Cys(430).
By determination of the number of surface-exposed cysteine residues and the number of persulfide-binding cysteine residues in combination with the sequential substitution of each of the nine cysteine residues, a second persulfide-binding cysteine residue, Cys(206), was identified.
Furthermore, the active-site Cys(430) was found to be located on top of a loop structure, formed by the two flanking residues Cys(428) and Cys(435), which are likely to form an intramolecular disulfide bridge.
These findings are confirmed by a structural model of the NifS-like domain, which indicates that Cys(428) and Cys(435) are within disulfide bond distance and that a persulfide transfer from Cys(430) to Cys(206) is indeed possible.
[‘NifS-like proteins are ubiquitous, homodimeric, proteins which belong to the α-family of pyridoxal-5’-phosphate dependent enzymes. They are proposed to donate elementary sulphur, generated from cysteine, via a cysteine persulphide intermediate during iron sulphur cluster biosynthesis.’ Kaiser, J.T., Clausen, T. Gleb P. Bourenkow, G.P., Bartunik, H.D., Steinbacher,S., and Huber, R.,J. Mol. Biol.,2000, 297, 451-464. Crystal Structure of a NifS-like Protein fromThermotoga maritima: Implications for Iron Sulphur Cluster Assembly.]
Lehrke, M., Rump, S., Heidenreich, T., Wissing, J., Mendel, R., Bittner, F., biochemical Journal, 2012, 441, 823-832. Identification of persulfide-binding and disulfide-forming cysteine residues in the NifS-like domain of the molybdenum cofactor sulfurase ABA3 by cysteine-scanning mutagenesis.
Biosynthesis reactions from Caspi et al, Nucleic Acids Research 38:D473-D479 2010 Page generated by SRI International Pathway Tools version 16.0 www.metacyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-5963&detail-level=3 ©2011 SRI International.
Molybdenum and enzymes molybdenum cofactor
Cyclic pyranopterin monophosphate
Hydrogenated pterins are found in all living organisms, where they are involved in key metabolic processes. Molybdenum in its biologically active form is bound to a fully reduced tetrahydropyranopterin, a metal-binding pterin (MPT), forming the molybdenum cofactor (Moco).
Cyclic pyranopterin monophosphate (cPMP) is the first isolatable intermediate in molybdenum cofactor biosynthesis. The (13)C NMR data for cPMP confirm the tetrahydropyranopterin nature of cPMP and the presence of a gem-diol in the C1' position of the side chain. The gem-diol is not a chemical artifact, but is chemically stable and not in equilibrium with the keto form.
The kinetics of cPMP oxidation in the presence of metal centers, chelating agents, and different buffers and pH values were studied. Oxidation is metal-dependent and can be retarded by EDTA.
Santamaria-Araujo, J.A., WrayV., Schwarz, G., Journal of Biological Inorganic Chemistry, 2012, 17, 1,113-122 Structure and stability of the molybdenum cofactor intermediate cyclic pyranopterin monophosphate.
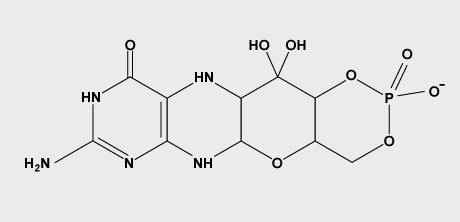
Cyclic pyranopterin monophosphate (cPMP)


